DNA Footprints: Using Parasites to Detect Elusive Animals, Proof of Principle in Hedgehogs
Abstract
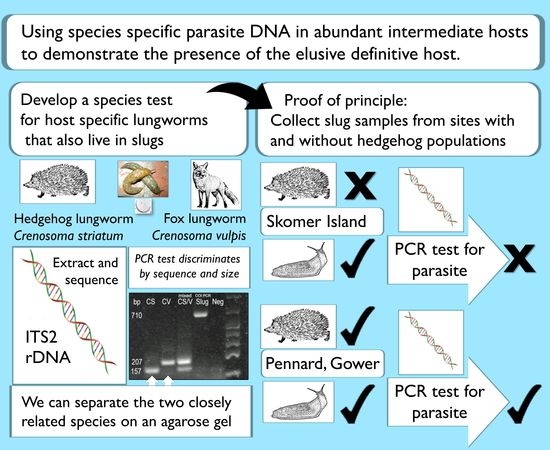
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of DNA from Nematodes for Molecular Test Development
2.2. Primer Design and Multiplex Assay Development
2.3. Slug Samples
2.4. Gastropod DNA Extraction
2.5. Sample Size Calculator
3. Results
4. Discussion
| Parasite Group | Helminth Species | Definitive Host | Common Name | Host Class | Reference |
|---|---|---|---|---|---|
| Nematode | |||||
| Abbreviata perenticola | Varanus giganteus | Perentie | Reptilia | [61] | |
| Abbreviata physignathi | Physignathus lesueurii | Australian Water Dragon | Reptilia | [61] | |
| Abbreviata glebopalmae | Varanus glebopalma | Black Palmed Rock Monitor | Reptilia | [61] | |
| Abbreviata barrowi | Pseudechis australis | Mulga Snake | Reptilia | [61] | |
| Nematodirus davtiani alpinus * | Capra ibex | Alpine Ibex | Mammalia | [62] | |
| Filaroides martes | Martes americana | American Pine Marten | Mammalia | [63] | |
| Perostrongylus falciformis | Meles meles | European badger | Mammalia | [64] | |
| Crenosoma striatum * | Erinaceus europaeus | European Hedgehog | Mammalia | ||
| Rhabdias bakeri | Rana sylvatica | Wood Frog | Amphibia | [57] | |
| Rhabdias ambystomae | Ambystoma maculatum | Spotted Salamander | Amphibia | [65] | |
| Parachordatortilis mathevossianae | Falco tinnunculus | Common Kestrel | Aves | [66] | |
| Physaloptera apivori | Pernis apivorus | European Honey Buzzard | Aves | [66] | |
| Physaloptera Mexicana * | Buteo buteo | Common Buzzard | Aves | [66] | |
| Serratospiculum tendo | Falco peregrinus | Peregrine falcon | Aves | [66] | |
| Trematode | |||||
| Urotrema scabridum * | Anolis sagrei | Brown Anole | Reptilia | [67] | |
| Pleurogonius malaclemys | Malaclemys terrapin | Diamondback Terrapin | Reptilia | [22] | |
| Mesocoelium lanfrediae | Rhinella marina | Cane Toad | Amphibia | [68] | |
| Parastrigea intermedia | Circus aeruginosus | Western Marsh Harrier | Aves | [66] | |
| Cestode | |||||
| Schistotaenia tenuicirrus | Podiceps grisegena | Red Necked Grebe | Aves | [69] | |
| Cladotaenia foxi | Falco Peregrinus | Peregrine falcon | Aves | [66] |
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| rDNA | ribosomal deoxyribonucleic acid |
| PCR | Polymerase chain reaction |
| dNTPs | deoxyribonucleotide triphosphate |
| mM | millimolar |
| bp | base pair |
| agg | aggregate |
| ha | hectare |
References
- Thompson, W.L. Sampling Rare or Elusive Species: Concepts, Designs and Techniques for Estimating Population Parameters; Island Press: Washington, DC, USA, 2004. [Google Scholar]
- Kendall, K.C.; Metzgar, L.H.; Patterson, D.A.; Steele, B.M. Power of Sign Surveys To Monitor Population Trends. Ecol. Appl. 1992, 2, 422–430. [Google Scholar] [CrossRef]
- Linnell, J.D.C.; Swenson, J.E.; Kvam, T.; Nikus, N.; Fagrapport, N.; Oppdragsmelding, N. Methods for Monitoring European Large Carnivores—A Worldwide Review of Relevant Experience; Institutt Norsk for Naturforskning: Trondheim, Norway, 1998; Volume 549, ISBN 8242609519. [Google Scholar]
- Eggert, L.S.; Eggert, J.A.; Woodruff, D.S. Estimating population sizes for elusive animals: The forest elephants of Kakum National Park, Ghana. Mol. Ecol. 2003, 12, 1389–1402. [Google Scholar] [CrossRef] [PubMed]
- Amori, G. Erinaceus Europaeus The IUCN Red List of Threatened Species 2016: E.T29650A2791303; IUCN: Gland, Switzerland, 2016. [Google Scholar] [CrossRef]
- Holsbeek, L.; Rodts, J.; Muyldermans, S. Hedgehog and other animal traffic victims in Belgium: Results of a countrywide survey. Lutra 1999, 42, 111–119. [Google Scholar]
- Huijser, M.P.; Bergers, P.J.M. The effect of roads and traffic on hedgehog (Erinaceus europaeus) populations. Biol. Conserv. 2000, 95, 111–116. [Google Scholar] [CrossRef]
- Macdonald, D.; Burnham, D. The State of Britains Mammals—People’s Trust for Endangered Species; The Wildlife Conservation Research Unit, Oxford: London, UK, 2011. [Google Scholar]
- Roos, S.; Johnston, A.; Noble, D. UK Hedgehog Datasets and Their Potential for Long-Term Monitoring; British Trust for Ornithology: Setford, UK, 2012; pp. 1–63. [Google Scholar]
- Krange, M. Change in the Occurrence of the West European Hedgehog (Erinaceus Europaeus) in Western Sweden during 1950–2010. Ph.D. Thesis, Karlstad University, Karlstad, Sweden, 2015. [Google Scholar]
- Hof, A.R.; Bright, P.W. Quantifying the long-term decline of the West European hedgehog in England by subsampling citizen-science datasets. Eur. J. Wildl. Res. 2016, 62, 407–413. [Google Scholar] [CrossRef]
- Wembridge, D. The State of Britain’s Hedgehogs 2011; British Hedgehog Preservation Society & Peoples Trust for Endangered Species: Ludlow, UK, 2011. [Google Scholar]
- Cavallini, P. Faeces count as an index of fox abundance. Acta 1994, 39, 417–424. [Google Scholar] [CrossRef] [Green Version]
- Silveira, L.; Jácomo, A.T.A.; Diniz-Filho, J.A.F. Camera trap, line transect census and track surveys: A comparative evaluation. Biol. Conserv. 2003, 114, 351–355. [Google Scholar] [CrossRef]
- McKelvey, K.S.; Kienast, J.V.; Aubry, K.B.; Koehler, G.M.; Maletzke, B.T.; Squires, J.R.; Linduist, E.L.; Loch, S.; Schwartz, M.K. DNA Analysis of Hair and Scat Collected Along Snow Tracks to Document the Presence of Canada Lynx. Wildl. Soc. Bull. 2006, 34, 451–455. [Google Scholar] [CrossRef]
- Yarnell, R.W.; Pacheco, M.; Williams, B.; Neumann, J.L.; Rymer, D.J.; Baker, P.J. Using occupancy analysis to validate the use of footprint tunnels as a method for monitoring the hedgehog Erinaceus europaeus. Mammal Rev. 2014, 44, 234–238. [Google Scholar] [CrossRef]
- McKelvey, K.S.; Aubry, K.B.; Schwartz, M.K. Using Anecdotal Occurrence Data for Rare or Elusive Species: The Illusion of Reality and a Call for Evidentiary Standards. Bioscience 2008, 58, 549. [Google Scholar] [CrossRef] [Green Version]
- Dogiel, V.; Bychowsky, B. Parasites of fisheries of the Caspian Sea. Tr. Kompleks. Izucheniyu Kaspiiskogo Morya 1939, 7, 150. [Google Scholar]
- Herrington, W.C.; Bearse, H.; Firth, F.E. Observations on the life history, occurence, and distribution of the redfish parasite Sphyrion lumpium. U. S. Bur. Fish. Spec. Rep. 1939, 5, 1–18. [Google Scholar]
- MacKenzie, K. Parasites as indicators of host populations. Int. J. Parasitol. 1987, 17, 345–352. [Google Scholar] [CrossRef]
- Williams, H.H.; MacKenzie, K.; McCarthy, A.M. Parasites as biological indicators of the population biology, migrations, diet, and phylogenetics of fish. Rev. Fish Biol. Fish. 1992, 2, 144–176. [Google Scholar] [CrossRef]
- Byers, J.E.; Altman, I.; Grosse, A.M.; Huspeni, T.C.; Maerz, J.C. Using parasitic trematode larvae to quantify an elusive vertebrate host. Conserv. Biol. 2011, 25, 85–93. [Google Scholar] [CrossRef]
- Skrjabin, K.I.; Petrow, A.M. A description of the genus Crenosoma Molin, 1861 (Metastrongylidae, Nematota). Parasitology 1928, 20, 329–335. [Google Scholar] [CrossRef]
- Dougherty, E.C. A review of the genus Crenosoma Molin, 1861 (Nematoda: Trichostrongylidae); its history, taxonomy, adult morphology, and distribution. J. Helminthol. Soc. Wash. 1945, 12, 44–62. [Google Scholar]
- Lammler, G.; Saupe, E. Infection experiments with the lungworm of the hedgehog, Crenosoma striatum. Z. Parasitenkd. 1968, 31, 87–100. [Google Scholar]
- Barus, V.; Blazek, K. The life cycle and the pathogenicity of the nematode Crenosoma-striatum. Folia Parasitol. 1971, 18, 215–226. [Google Scholar]
- Reeve, N. Hedgehogs; T. & A.D. Poyser: London, UK, 1994; ISBN1 085661081X. ISBN2 9780856610813. [Google Scholar]
- Mirzaei, M. Infection with Crenosoma striatum lungworm in Long-eared Hedgehog (Hemiechinus auritus) in Kerman province southeast of Iran. Turk. J. Parasitol. 2015, 38, 255–257. [Google Scholar] [CrossRef]
- Gaglio, G.; Allen, S.; Bowden, L.; Bryant, M.; Morgan, E. Parasites of European hedgehogs (Erinaceus europaeus) in Britain: Epidemiological study and coprological test evaluation. Eur. J. Wildl. Res. 2010, 56, 839–844. [Google Scholar] [CrossRef]
- Grewal, P.S.; Grewal, S.K.; Tan, L.; Adams, B.J. Parasitism of molluscs by nematodes: Types of associations and evolutionary trends. J. Nematol. 2003, 35, 146–156. [Google Scholar] [PubMed]
- Mason, C.F. Snail populations, beech litter production, and role of snails in litter decomposition. Oecologia 1970, 5, 213–239. [Google Scholar] [CrossRef] [PubMed]
- South, A. Terrestrial Slugs, Biology, Ecology and Control; Chapman and Hall: London, UK, 1992; ISBN 0-412-36810-2. [Google Scholar]
- Kennedy, C.R. Ecology of the Acanthocephala; Cambridge University Press: Cambridge, UK, 2006; ISBN 9780521850087. [Google Scholar]
- Chilton, N.B.; Gasser, R.B.; Beveridge, I. Differences in a ribosomal DNA sequence of morphologically indistinguishable species within the Hypodontus macropi complex (Nematoda: Strongyloidea). Int. J. Parasitol. 1995, 25, 647–651. [Google Scholar] [CrossRef]
- Félix, M.A.; Braendle, C.; Cutter, A.D. A streamlined system for species diagnosis in caenorhabditis (Nematoda: Rhabditidae) with name designations for 15 distinct biological species. PLoS ONE 2014, 9, e0118327. [Google Scholar] [CrossRef]
- Gasser, R.B.; Monti, J.R. Identification of parasitic nematodes by PCR-SSCP of ITS-2 rDNA. Mol. Cell. Probes 1997, 11, 201–209. [Google Scholar] [CrossRef]
- Sinclair, R.; Melville, L.; Sargison, F.; Kenyon, F.; Nussey, D.; Watt, K.; Sargison, N. Gastrointestinal nematode species diversity in Soay sheep kept in a natural environment without active parasite control. Vet. Parasitol. 2016, 227, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Wimmer, B.; Craig, B.H.; Pilkington, J.G.; Pemberton, J.M. Non-invasive assessment of parasitic nematode species diversity in wild Soay sheep using molecular markers. Int. J. Parasitol. 2004, 34, 625–631. [Google Scholar] [CrossRef]
- Gasser, R.B. PCR-based technology in veterinary parasitology. Vet. Parasitol. 1999, 84, 229–258. [Google Scholar] [CrossRef]
- Gasser, R.B. Molecular tools—Advances, opportunities and prospects. Vet. Parasitol. 2006, 136, 69–89. [Google Scholar] [CrossRef]
- Gasser, R.B.; Chilton, N.B.; Hoste, H.; Beveridge, I. Rapid sequencing of rDNA from single worms and eggs of parasitic helminths. Nucleic Acids Res. 1993, 21, 2525–2526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 45, 95–98. [Google Scholar]
- Aziz, N.A.A.; Daly, E.; Allen, S.; Rowson, B.; Greig, C.; Forman, D.; Morgan, E.R. Distribution of Angiostrongylus vasorum and its gastropod intermediate hosts along the rural–urban gradient in two cities in the United Kingdom, using real time PCR. Parasit. Vectors 2016, 9, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Folmer, O.; Black, M.; Hoeh, W.; Lutz, R.; Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar] [PubMed]
- Hilborn, R.; Mangel, M. The Ecological Detective: Confronting models with data. In The Ecological Detective; Princeton University Press: Princeton, NJ, USA, 1997; p. 315. ISBN 978-0691034966. [Google Scholar]
- Piaggio, A.J.; Engeman, R.M.; Hopken, M.W.; Humphrey, J.S.; Keacher, K.L.; Bruce, W.E.; Avery, M.L. Detecting an elusive invasive species: A diagnostic PCR to detect Burmese python in Florida waters and an assessment of persistence of environmental DNA. Mol. Ecol. Resour. 2014, 14, 374–380. [Google Scholar] [CrossRef] [Green Version]
- Mills, L.S.; Citta, J.J.; Lair, K.P.; Schwartz, M.K.; Tallmon, D.A. Estimating Animal Abundance Using Noninvasive DNA Sampling: Promise and Pitfalls. Ecol. Appl. 2000, 10, 283–294. [Google Scholar] [CrossRef]
- Jones, R.A.; Brophy, P.M.; Davis, C.N.; Davies, T.E.; Emberson, H.; Stevens, P.R.; Williams, H.W. Detection of Galba truncatula, Fasciola hepatica and Calicophoron daubneyi environmental DNA within water sources on pasture land, a future tool for fluke control? Parasit. Vectors 2018, 11, 1–9. [Google Scholar] [CrossRef]
- Morgan, E.R.; Shaw, S.E.; Brennan, S.F.; De Waal, T.D.; Jones, B.R.; Mulcahy, G. Angiostrongylus vasorum: A real heartbreaker. Trends Parasitol. 2005, 21, 49–51. [Google Scholar] [CrossRef]
- Barnes, H.F.; Weil, J.W. Slugs in Gardens: Their Numbers, Activities and Distribution. Part 1. J. Anim. Ecol. 1944, 13, 140–175. [Google Scholar] [CrossRef]
- Poulin, R.; Keeney, D.B. Host specificity under molecular and experimental scrutiny. Trends Parasitol. 2007. [Google Scholar] [CrossRef]
- Desdevises, Y.; Morand, S.; Legendre, P. Evolution and determinants of host specificity in the genus Lamellodiscus ( Monogenea ). Biol. J. Linn. Soc. 2002, 77, 431–443. [Google Scholar] [CrossRef] [Green Version]
- Cleaveland, S.; Laurenson, M.K.; Taylor, L.H. Diseases of humans and their domestic mammals: Pathogen characteristics, host range and the risk of emergence. Philos. Trans. R. Soc. Lond. 2001, 356, 991–999. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, A.B.; Altizer, S.; Poss, M.; Cunningham, A.A.; Nunn, C.L. Patterns of host specificity and transmission among parasites of wild primates. Int. J. Parasitol. 2005, 35, 647–657. [Google Scholar] [CrossRef] [PubMed]
- Woolhouse, M.E.J.; Taylor, L.H.; Haydon, D.T. Population Biology of Multihost Pathogens. Science 2001, 1828, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Poulin, R.; Krasnov, B.R.; Mouillot, D. Host specificity in phylogenetic and geographic space. Trends Parasitol. 2011, 27, 355–361. [Google Scholar] [CrossRef]
- Tkach, V.V.; Kuzmin, Y.; Pulis, E.E. A New Species of Rhabdias From Lungs of the Wood Frog, Rana Sylvatica, in North America: The Last Sibling of Rhabdias Ranae? J. Parasitol. 2006, 92, 631–636. [Google Scholar] [CrossRef]
- Cable, J.; Barber, I.; Boag, B.; Ellison, A.R.; Morgan, E.R.; Murray, K.; Pascoe, E.L.; Sait, S.M.; Wilson, A.J.; Booth, M. Global change, parasite transmission and disease control: Lessons from ecology. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372. [Google Scholar] [CrossRef]
- Feng, K.; Li, W.; Guo, Z.; Duo, H.; Fu, Y.; Shen, X.; Tie, C.; Rijie, E.; Changqin, X.; Luo, Y.; et al. Development of LAMP assays for the molecular detection of taeniid infection in canine in Tibetan rural area. J. Vet. Med. Sci. 2017, 79, 1986–1993. [Google Scholar] [CrossRef] [Green Version]
- Abbasi, I.; King, C.H.; Muchiri, E.M.; Hamburger, J. Detection of Schistosoma mansoni and Schistosoma haematobium DNA by loop-mediated isothermal amplification: Identification of infected snails from early prepatency. Am. J. Trop. Med. Hyg. 2010, 83, 427–432. [Google Scholar] [CrossRef] [Green Version]
- Jones, H.I. Physalopterine nematodes in Australian reptiles: Interactions and patterns of infection. Aust. J. Zool. 2014, 62, 180–194. [Google Scholar] [CrossRef] [Green Version]
- Zaffaroni, E.; Manfredi, M.T.; Citterio, C.; Sala, M.; Piccolo, G.; Lanfranchi, P. Host specificity of abomasal nematodes in free ranging alpine ruminants. Vet. Parasitol. 2000, 90, 221–230. [Google Scholar] [CrossRef]
- Seville, R.S.; Addison, E.M. Nongastrointestinal helminths in marten (Martes americana) from Ontario, Canada. J. Wildl. Dis. 1995, 31, 529–533. [Google Scholar] [CrossRef] [PubMed]
- Deak, G.; Mihalca, A.D.; Hirzmann, J.; Colella, V.; Alexandru, F.; Cavalera, M.A.; Gheorghe, F.; Bauer, C.; Monica, A.; Ali, A.; et al. Validity of genus Perostrongylus Schlegel, 1934 with new data on Perostrongylus falciformis (Schlegel, 1933) in European badgers, Meles meles (Linnaeus, 1758): Distribution, life-cycle and pathology. Parasite Vector 2018, 56, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Kuzmin, Y.; Tkach, V.V.; Snyder, S.D. Rhabdias ambystomae sp. n. (Nematoda: Rhabdiasidae) from the north American spotted salamander Ambystoma maculatum (Amphibia: Ambystomatidae). Comp. Parasitol. 2001, 68, 228–235. [Google Scholar]
- Santoro, M.; Kinsella, J.M.; Galiero, G.; degli Uberti, B.; Aznar, F.J. Helminth Community Structure in Birds of Prey (Accipitriformes and Falconiformes) in Southern Italy. J. Parasitol. 2012, 98, 22–29. [Google Scholar] [CrossRef]
- Bundy, D.A.P.; Harris, E.A. Helminth parasites of jamaican anoles (Reptilia: Iguanidae): A comparison of the helminth fauna of 6 anolis species. J. Helminthol. 1987, 61, 77–83. [Google Scholar] [CrossRef]
- Gomes, T.F.F.; Melo, F.T.V.; Giese, E.G.; Furtado, A.P.; Gonçalves, E.C.; Santos, J.N. A new species of Mesocoelium (Digenea: Mesocoeliidae) found in Rhinella marina (Amphibia: Bufonidae) from Brazilian Amazonia. Mem. Inst. Oswaldo Cruz 2013, 108, 186–191. [Google Scholar] [CrossRef] [Green Version]
- Stock, T.M.; Holmes, J.C. Host specificity and exchange of intestinal helminths among four species of grebes (Podicipedidae). Can. J. Zool. 1987, 65, 669–676. [Google Scholar] [CrossRef]

| Primer | Sequence (5′-3′) | Locus | References |
|---|---|---|---|
| NC1 | ACGTCTGGTTCAGGGTTGTT | 5.8S nuclear rRNA gene | [41] |
| NC2 | TTAGTTTCTTTTCCTCCGCT | 28S nuclear rRNA gene | [41] |
| C.St-ITS2F | CGATTCCCGTTCTAGTTGAGAC | ITS-2 nuclear rDNA | this study |
| C.St-ITS2R | AAAACCACCTCGACGACATC | ITS-2 nuclear rDNA | this study |
| C.Vu-ITS2F | CGATTCCCGTTTTAGTTAAGGA | ITS-2 nuclear rDNA | this study |
| C.Vu-ITS2R | GCTTATCAATCGTCGAATATCATGC | ITS-2 nuclear rDNA | this study |
| LCO1490 | GGTCAACAAATCATAAAGATATTGG | Mitochondrial cox1 gene | [44] |
| HC02198 | TAAACTTCAGGGTGACCAAAAAATCA | Mitochondrial cox1 gene | [44] |
| DNA | 260/280 | Weight | ||||
| Sample ID | Slug Species | μg/mL | Ratio | mg | COI | Excluded |
| Pennard Spring | ||||||
| 1 | Tandonia sowerbyi | 59.12 | 1.54 | 50 | +ve | |
| 2 | Tandonia sowerbyi | 86.36 | 1.60 | 34 | +ve | |
| 3 | Tandonia sowerbyi | 45.64 | 1.60 | 40 | +ve | |
| 4 | Tandonia sowerbyi | 102.74 | 1.83 | 47 | −ve | x |
| 5 | Deroceras panormitanum | 151.01 | 1.91 | 42 | −ve | x |
| 6 | Deroceras panormitanum | 179.24 | 2.00 | 63 | +ve | |
| 7 | Deroceras panormitanum | 180.09 | 1.97 | 55 | −ve | x |
| 8 | Deroceras panormitanum | 94.04 | 1.79 | 48 | −ve | x |
| 9 | Lehmannia marginata | 7.19 | 1.37 | 50 | +ve | |
| 10 | Lehmannia marginata | 13.98 | 1.42 | 56 | +ve | |
| 11 | Lehmannia marginata | 108.34 | 1.77 | 42 | −ve | x |
| 12 | Lehmannia marginata | 54.50 | 1.89 | 48 | +ve | |
| 13 | Arion hortensis agg. | 30.28 | 1.55 | 45 | +ve | |
| 14 | Arion hortensis agg. | 37.04 | 1.70 | 41 | +ve | |
| 15 | Arion hortensis agg. | 25.47 | 1.63 | 38 | +ve | |
| 16 | Arion hortensis agg. | 26.16 | 1.49 | 50 | −ve | x |
| SL-1 | Tandonia sowerbyi | 32.00 | 2.05 | 200 | +ve | |
| SL-2 | Tandonia sowerbyi | 325.69 | 2.03 | 141 | +ve | |
| SL-3 | Tandonia sowerbyi | 310.11 | 2.04 | +ve | ||
| SL-4 | Arion subfuscus | 464.97 | 2.00 | +ve | ||
| SL-5 | Arion subfuscus | 300.12 | 2.04 | −ve | x | |
| SL-6 | Arion subfuscus | 73.10 | 1.84 | −ve | x | |
| SL-7 | Arion subfuscus | 173.21 | 1.98 | −ve | x | |
| SL-8 | Arion subfuscus | 128.40 | 1.94 | −ve | x | |
| SL-9 | Arion flagellus | 45.35 | 1.89 | +ve | ||
| SL-10 | Arion flagellus | 226.78 | 2.02 | +ve | ||
| SL-11 | Arion flagellus | 153.25 | 1.90 | +ve | ||
| SL-12 | Arion flagellus | 107.87 | 1.89 | +ve | ||
| SL-13 | Arion flagellus | 107.57 | 1.89 | +ve | ||
| SL-14 | Arion flagellus | 200.06 | 1.99 | +ve | ||
| Pennard Autumn | ||||||
| 11.1 | Arion ater agg. | 27.62 | 1.46 | 60 | +ve | |
| 11.2 | Arion ater agg. | 46.00 | 1.81 | 50 | +ve | |
| 11.3 | Arion ater agg. | 23.00 | 1.61 | 50 | +ve | |
| 11.4 | Arion ater agg. | 57.00 | 1.71 | 50 | +ve | |
| 28.1 | Arion ater agg. | 107.77 | 1.70 | 50 | −ve | x |
| 28.2 | Arion ater agg. | 44.54 | 1.68 | 50 | +ve | |
| 28.3 | Arion ater agg. | 31.04 | 1.41 | 50 | −ve | x |
| 28.4 | Arion ater agg. | 44.49 | 1.61 | 50 | +ve | |
| 15.1 | Arion ater agg. | 32.01 | 1.49 | 50 | +ve | |
| 15.2 | Arion ater agg. | 62.77 | 1.67 | 50 | +ve | |
| 15.3 | Arion ater agg. | 10.13 | 1.12 | 50 | +ve | |
| 15.4 | Arion ater agg. | 61.56 | 1.80 | 80 | +ve | |
| 2.1 | Limax flavus | 0.00 | −0.69 | 50 | −ve | x |
| 2.2 | Limax flavus | 17.91 | 1.26 | 50 | +ve | |
| 2.3 | Limax flavus | 6.27 | 1.12 | 60 | +ve | |
| 2.4 | Limax flavus | 93.23 | 1.77 | 60 | +ve | |
| 1.1 | Tandonia sowerbyi | 76.78 | 0.92 | 58 | +ve | |
| 1.2 | Tandonia sowerbyi | 43.57 | 1.57 | 54 | +ve | |
| 1.3 | Tandonia sowerbyi | 56.92 | 1.70 | 44 | +ve | |
| 1.4 | Tandonia sowerbyi | 40.62 | 1.49 | 47 | +ve | |
| 22.1 | Arion rufus | 2.55 | 0.72 | 58 | +ve | |
| 22.2 | Arion rufus | 0.59 | 0.61 | 48 | −ve | x |
| 22.3 | Arion rufus | 19.40 | 1.21 | 41 | +ve | |
| 22.4 | Arion rufus | 11.28 | 1.06 | 52 | +ve | |
| 23.1 | Arion rufus | 7.08 | 1.08 | 49 | +ve | |
| 23.2 | Arion rufus | 25.60 | 1.34 | 57 | +ve | |
| Skomer Autumn | ||||||
| SK1 | Arion ater | 179.7 | 1.8 | 30 | +ve | |
| SK2 | Arion ater | 114 | 1.85 | +ve | ||
| SK3 | Arion ater | 53.2 | 1.87 | +ve | ||
| SK4 | Arion ater | 105.1 | 1.83 | +ve | ||
| SK5 | Arion ater | 129.1 | 1.85 | 60 | +ve | |
| SK6 | Arion ater | 172.3 | 1.75 | +ve | ||
| SK7 | Arion ater | 160.3 | 1.64 | 100 | −ve | x |
| SK8 | Arion ater | 271.4 | 1.97 | 20 | −ve | x |
| SK9 | Arion ater | 266.8 | 1.93 | 41 | −ve | x |
| SK10 | Arion ater | 118.8 | 1.86 | +ve | ||
| SK11 | Lehmannia marginata | 107.4 | 1.86 | 48 | +ve | |
| SK12 | Lehmannia marginata | 150.8 | 1.95 | +ve | ||
| SK13 | Lehmannia marginata | 174.7 | 1.93 | 19 | +ve | |
| SK14 | Lehmannia marginata | 166.4 | 2.02 | +ve | ||
| SK15 | Lehmannia marginata | 161.3 | 1.93 | +ve | ||
| SK16 | Lehmannia marginata | 279.7 | 1.95 | +ve | ||
| SK17 | Lehmannia marginata | 355.2 | 1.95 | +ve | ||
| SK18 | Arion ater | 100.1 | 1.91 | +ve | ||
| SK19 | Arion ater | 73.1 | 1.82 | +ve | ||
| SK20 | Arion ater | 97.2 | 1.87 | 83 | +ve | |
| SK21 | Arion ater | 108.7 | 2.1 | +ve | ||
| SK22 | Arion ater | 233.4 | 1.92 | +ve | ||
| SK23 | Arion ater | 130.9 | 1.99 | +ve | ||
| SK24 | Arion ater | 130.6 | 1.9 | +ve | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Allen, S.; Greig, C.; Rowson, B.; Gasser, R.B.; Jabbar, A.; Morelli, S.; Morgan, E.R.; Wood, M.; Forman, D. DNA Footprints: Using Parasites to Detect Elusive Animals, Proof of Principle in Hedgehogs. Animals 2020, 10, 1420. https://doi.org/10.3390/ani10081420
Allen S, Greig C, Rowson B, Gasser RB, Jabbar A, Morelli S, Morgan ER, Wood M, Forman D. DNA Footprints: Using Parasites to Detect Elusive Animals, Proof of Principle in Hedgehogs. Animals. 2020; 10(8):1420. https://doi.org/10.3390/ani10081420
Chicago/Turabian StyleAllen, Simon, Carolyn Greig, Ben Rowson, Robin B. Gasser, Abdul Jabbar, Simone Morelli, Eric R. Morgan, Martyn Wood, and Dan Forman. 2020. "DNA Footprints: Using Parasites to Detect Elusive Animals, Proof of Principle in Hedgehogs" Animals 10, no. 8: 1420. https://doi.org/10.3390/ani10081420